THE BOBO-DIOULASSO PLATFORM










Laboratoire de Bobo-Dioulasso
Operational platforms
- Molecular biology platform :
DNA extractions, PCR amplification and visualization
– Detection of major bacterial and fungal pathogens
– Sequencing targetted genes
– Experimental infections in semi controlled conditions
- Bacteriology platform
- Mycology platform
- Nematology platform
Diagnostic bactériens, champignons, nématodes, infections expérimentales en conditions semi-contrôlées


The molecular laboratory (former PV site) where DNA extractions and amplification by PCR are carried out. Biology
- Multi disciplinary study plots on rice
Developed each year since 2014 on two sites (lowlands of Banfora and irrigated perimeters of Karfiguela), these experimental plots situated within the agro-ecosystem enabling to conduct a pluri-annual monitory of the set of bio-aggressors (virus, bacteria, fungi, nematodes, insects and self-propagating plants) thanks to a collaborative work associating phytopathology, entomology and weed science.

Picture of the experimental plot of Banfora : four cultivars certified in Burkina Faso are placed in random blocks with weeding and herb planting treatment.
Research focus

- Genetic structure of pathogen populations
The structure of pathogen populations is studied with microsatellite markers for various Xanthomonas species (affecting rice, cassava or mango trees) or fungi, such as blast disease in rice.
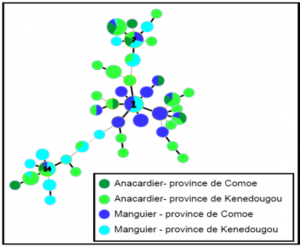 Results show no genetic difference between the Xanthomonas citri pv. Mangiferaeindicae populations collected on a mango tree and a cashew tree.
Results show no genetic difference between the Xanthomonas citri pv. Mangiferaeindicae populations collected on a mango tree and a cashew tree.
The pathogenic data are in line with these genetic data, enabling to conclude on lack of host specialization with this bacteria species.
Zombre et al. (2016)
- Control method: cultivar resistance, plant extract
 Results gathered in controlled conditions have shown that rice cultivars certified in Burkina Faso are resistant to Xanthomonas oryzae.
Results gathered in controlled conditions have shown that rice cultivars certified in Burkina Faso are resistant to Xanthomonas oryzae.
Field tests are ongoing in order to assess the sustainability of such resistances and to detect possible resistance breakdown events.
 The use of plant extracts is another control method that has been studied. For example, the essential oil of Cymbopogon citratus inhibits the growth of Xanthomonas oryzae in vitro.
The use of plant extracts is another control method that has been studied. For example, the essential oil of Cymbopogon citratus inhibits the growth of Xanthomonas oryzae in vitro.
Wonni et al. (2016)
- Studying co-infection and multi-pathogen epidemiological monitories
 The results obtained on the interaction between the RYMV virus and Xoc bacteria in rice reveal that co-infection is frequent in the agro-ecosystem. Experimentally, a reciprocal effect was highlighted in a co-infection context, likely involving “RNA silencing” plant defense mechanism.
The results obtained on the interaction between the RYMV virus and Xoc bacteria in rice reveal that co-infection is frequent in the agro-ecosystem. Experimentally, a reciprocal effect was highlighted in a co-infection context, likely involving “RNA silencing” plant defense mechanism.
Tollenaere et al (2017)
Multiple disease epidemiological surveys (see: https://dataverse.ird.fr/dataset.xhtml?persistentId=doi:10.23708/8FDWIE ) and experimental approaches are carried out (see EVCOPAR project) in order to study the interactions between the different major rice pathogens.
- Plant associated microbiota
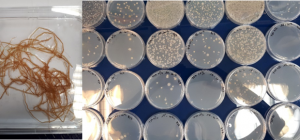 The characterization bacterial and fungal communities associated with plants is performed by targeted detection, metabarcoding and microbiology approaches. The last approach (“culturomics”) enables to consider in vivo tests of micro-organisms studied to assess their potential bio-fertilizing and/or biocontrol properties.
The characterization bacterial and fungal communities associated with plants is performed by targeted detection, metabarcoding and microbiology approaches. The last approach (“culturomics”) enables to consider in vivo tests of micro-organisms studied to assess their potential bio-fertilizing and/or biocontrol properties.
Training
- Supervision of from many universities and centers in Burkina Faso (Nazi Boni University, Versatile Agricultural Center of Matourkou, University Ouaga 1 Joseph Ki-Zerbo, New Dawn University) or abroad (Cheikh Anta Diop University, Felix Houphouet-Boigny University)
- Organisation of specific workshops on various thematic areas such as molecular detection, sequencing, data analysis and bioinformatics, in collaboration with universities in Burkina Faso
- Capacity building for agricultural farmers and agents on agricultural practices

Top left and in center: a training on molecular biology for students (September 2016). Bottom left: a workshop on “Strategies for innovative control of bacterial diseases, based on knowledge on molecular mechanisms of interaction” (October 2017). On the right (top and bottom): a training on seed production for rice farmers.
Latest publications
- Kassankogno A, Ouedrogo I, Tiendrebeogo A, Ouedraogo L, Sankara P. 2015. In vitro evaluation of the effect of aqueous extracts of Agave sisalana and Cymbopogon citratus on mycelial growth and conidia production of Pyricularia oryzae, causal agent of rice blast. Journal of Applied Biosciences. 89
- Tollenaere C, Lacombe S, Wonni I, Barro M, Ndougonna C, Gnacko F, Sérémé D, Jacobs J, Hebrard E, Cunnac S, Brugidou C. 2017. Virus-bacteria rice co-infection in Africa: field estimation, reciprocal effects, molecular mechanisms, and evolutionary implications. Frontiers in Plant Science. 8:645.
- Wonni I, Cottyn B, Detemmerman L, Dao S, Ouedraogo L, Sarra S, Tekete C, Poussier S, Corral R, Triplett L, Koita O, Koebnik R, Leach J, Szurek B, Maes M, Verdier V. 2014. Analysis of Xanthomonas oryzae pv. oryzicola population in Mali and Burkina Faso reveals a high level of genetic and pathogenic diversity. Phytopathology. 104:520-31.
- Wonni I, Hutin M, Ouédrago L, Somda I, Verdier V. 2016. Evaluation of elite rice varieties unmasks new sources of bacterial blight and leaf streak resistance for Africa. Journal of Rice Research. 4: 162.
- Wonni I, Ouedraogo SL, Ouedraogo I, Sanogo L. 2016. Antibacterial activity of extracts of three aromaticplants from Burkina Faso against rice pathogen, Xanthomanas oryzae. African Journal of Microbiology. 10: 681-686.
- Zombre C, Sankara P, Ouédraogo SL, Wonni I, Boyer K, Boyer C, Terville M, Javegny S, Allibert A, Vernière C, Pruvost O. 2016. Natural Infection of cashew (Anacardium occidentale) by Xanthomonas citri pv. mangiferaeindicae in Burkina Faso. Plant Disease, 100 (4): 718-723.
